Genetic analysis using whole-exome sequencing in pediatric chronic kidney disease: a single center's experience
Article information
Abstract
Purpose
Chronic kidney disease (CKD) has various underlying causes in children. Identification of the underlying causes of CKD is important. Genetic causes comprise a significant proportion of pediatric CKD cases.
Methods
In this study, we performed whole-exome sequencing (WES) to identify genetic causes of pediatric CKD. From January to June 2021, WES was performed using samples from pediatric patients with CKD of unclear etiology.
Results
Genetic causes were investigated using WES in 37 patients (17 males) with pediatric CKD stages 1 (n=5), 2 (n=7), 3 (n=2), 4 (n=2), and 5 (n=21). The underlying diseases were focal segmental glomerulosclerosis (n=9), congenital anomalies of the kidney and urinary tract including reflux nephropathy (n=8), other glomerulopathies (n=7), unknown etiology (n=6), and others (n=7). WES identified genetic causes of CKD in 12 of the 37 patients (32.4%). Genetic defects were discovered in the COL4A4 (n=2), WT1 (n=2), ACTN4, CEP290, COL4A3, CUBN, GATA3, LAMA5, NUP107, and PAX2 genes. WT1 defects were found in patients whose pathologic diagnosis was membranoproliferative glomerulonephritis, and identification of CUBN defects led to discontinuation of immunosuppressive agents. Genetic diagnosis confirmed the clinical diagnosis of hypoparathyroidism, sensorineural deafness, and renal disease; Alport syndrome; and Joubert syndrome in three of the patients with CKD of unknown etiology (COL4A4 [n=2], CUBN [n=1]). Extrarenal symptoms were considered phenotypic presentations of WT1, PAX2, and CEP290 defects.
Conclusions
WES provided a genetic diagnosis that confirmed the clinical diagnosis in a significant proportion (32.4%) of patients with pediatric CKD.
Introduction
Chronic kidney disease (CKD) is a global health problem with increasing incidence and prevalence [1]. Children with CKD face mortality, lifelong morbidity, and a low quality of life [2-5]. Advancements in medical care have substantially improved the survival rate of children with CKD [1]. Identifying the underlying cause of this disease is essential because the progression, treatment, and prognosis may differ according to etiology. However, traditional diagnostic approaches, such as kidney biopsy, could be unrevealing or contraindicated when the kidneys are already failing [6]. Therefore, new diagnostic methods are required to identify the etiology of CKD.
CKD is a complex genetically heterogeneous disease with both genomic and environmental causes. The heritability of CKD is relatively high (30%–75%) [7], and at least 15% to 20% of early onset CKD (before the age of 25 years) is caused by genetic variation. Nearly all children who progress to end-stage kidney disease (ESKD) have an inherited form of CKD [8,9]. In addition, approximately 17% of patients with ESKD do not have a primary renal disease diagnosis and are therefore labeled as patients with CKD of unknown etiology.
The diagnostic accuracy provided by genetic testing [6] could enable the establishment of treatment guidelines and aid in the accurate prediction of patient prognosis. Genetic diagnosis is crucial for identifying high-risk groups and for appropriate family planning. The prevalence of CKD caused by genetic defects is approximately 10% in unselected adults [9] and 20% to 30% in children with nephropathy [7,10]. These findings indicate that the clinical application of genetic testing could transform diagnostic pathways by providing a timely and accurate genetic diagnosis [11].
Sanger sequencing of the causative genes is typically performed to obtain a genetic diagnosis when CKD is suspected. However, the number of known causative genes for CKD is increasing, as is the number of CKD cases. Therefore, traditional Sanger sequencing is not cost-effective in most cases, making targeted exome sequencing or whole-exome sequencing (WES) the preferred approaches for genetic diagnosis [12-15]. WES can screen most genes associated with diseases and can therefore be applied across diverse categories of renal disorders. In addition, it can potentially identify novel etiological genes associated with nephropathy. Therefore, WES is emerging as the preferred diagnostic tool for hereditary disorders [12-17]. It has provided a genetic diagnosis in up to 11.5%, 26%, and 32.7% of patients with congenital kidney anomalies, steroid-resistant nephrotic syndrome, and ESKD, respectively [18,19].
In this study, we present the preliminary results of utilizing WES to identify the genetic causes of pediatric CKD in Republic of Korea.
Methods
1. Study design and participants
We prospectively recruited 37 pediatric patients with CKD whose underlying etiology was uncertain or suspected to be monogenic. The male to female ratio of the patients was 20:17. All study participants were recruited from Seoul National University Children’s Hospital, Seoul, Republic of Korea. Buccal mucosal samples were collected and analyzed from January to June 2021. Details of DNA extraction and analysis of WES data have been previously described [20]. The identified variants were classified based on the American College of Medical Genetics and Genomics standards for the interpretation of sequence variants [21].
2. Statistical analysis
The diagnostic yield was calculated based on the variants classified as “pathogenic” or “likely pathogenic.” All statistical analyses were conducted using Microsoft Excel (Microsoft Corp., Redmond, WA, USA) and SPSS statistical software version 20 (IBM Corp, Armonk, NY, USA).
Results
1. Cohort characteristics
A total of 37 patients with CKD were recruited for this study. The underlying causes of CKD among the study participants were focal segmental glomerulosclerosis, congenital anomalies of the kidney and urinary tract (CAKUT) including reflux nephropathy, ischemic disease, Fanconi syndrome, Bartter syndrome, and CKD of unknown etiology (Table 1). One patient had a family history of CKD. The median ages at the time of CKD diagnosis and recruitment were 6 and 13 years, respectively. The distributions of the CKD stages and clinical diagnoses are described in Tables 1 and 2.
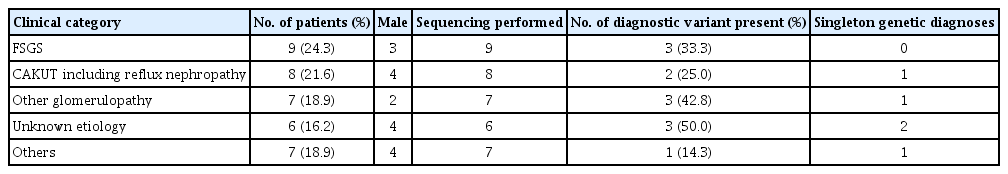
Clinical category characteristic and diagnostic yield of genetic diagnose across clinical diagnostic categories
2. Genetic findings and diagnostic yield
Diagnostic variants were detected in 12 of the 37 patients (32.4%), encompassing seven distinct monogenic disorders. The detected genes were COL4A4 (n=2), WT1 (n=2), ACTN4 (n=1), CEP290 (n=1), COL4A3 (n=1), CUBN (n=1), GATA3 (n=1), LAMA5 (n=1), NUP107 (n=1), and PAX2 (n=1). An additional three patients had variants of uncertain significance. The diagnostic yield (Table 1) was the highest among patients with CKD of unknown etiology (n=6, 50%). In contrast, diagnostic variants were not detected in some (n=7) of the clinically diagnosed CKD cases, including Alport syndrome, CAKUT, and renal hypoplasia with diabetes mellitus.
3. Clinical implications of genetic diagnoses in the study
Table 3 summarizes the clinical characteristics and genetic results of the patients with diagnostic variants. In six cases including three patients with CKD of unknown etiology, WES clarified the clinical diagnosis or reclassified the disease, which considerably affected clinical decision making. In five cases, WES confirmed the clinical diagnosis of hypoparathyroidism, sensorineural deafness, and renal disease; Alport syndrome; and Joubert syndrome.
Genetic diagnoses have direct consequences for medical management. The diagnosis of Alport syndrome enabled genetic counseling and screening for auditory and ophthalmological problems. Detection of WT1 defects in patients with pathological diagnoses of membranoproliferative glomerulonephritis led to more careful monitoring of malignancy. In accordance with Kidney Disease: Improving Global Outcomes (KDIGO) guidelines, identification of CUBN defects in a patient with persistent isolated proteinuria resulted in discontinuation of immunosuppressive agents [22]. Patients with extrarenal symptoms were examined to obtain a genetic diagnosis of WT1 (sex reversal), PAX2 (retinopathy), and CEP290 (Joubert syndrome) defects.
Discussion
In this study, we used WES to identify the genetic causes of pediatric CKD in Republic of Korea. The diagnostic yield of this approach was 32.4%, encompassing 10 genes, one copy number variation, and one microdeletion. This is similar to previous studies reporting detection rates of causal mutations. Mutations were detected in approximately 20% and 30% of patients who presented with CKD [7] and ESKD [10], respectively. These causal mutations were detected in patients diagnosed before the age of 25 years. Furthermore, children with kidney failure had a detection rate of approximately 40% [6]. With rapid technological advances, we expect that regular re-analysis, re-interpretation, and reclassification of variants will increase these detection rates [23,24].
The primary diagnosis in patients with ESKD is often inaccurate [25], resulting in its characterization as a CKD of unknown origin. In adults, WES provided a genetic diagnosis in 22 out of 92 patients (24%) with CKD of unknown etiology [26], whereas our study established a diagnostic yield of 50%. This shows that WES is effective in determining the genetic causes of CKD of unknown etiology. However, it is important to note that the number of patients in our study was small.
Establishing a precise genetic diagnosis can allow for the preemptive screening of extrarenal manifestations. Patients clinically diagnosed with isolated CAKUT can have mutations in genes that cause syndromic diseases [27]. In other cases, extrarenal manifestations could be detected later in life. Moreover, subtle phenotypes may be initially overlooked and only identified through a genetic diagnosis. In these cases of reverse phenotyping, identification of a genetic mutation can lead to preemptive screening for extrarenal manifestations, leading to early provision of treatment where possible. In our study, extrarenal symptoms were identified to obtain a genetic diagnosis of WT1, PAX2, and CEP290 defects.
Recent KDIGO guidelines recommend discontinuing immunosuppressive agents if a monogenic cause is discovered in a patient with focal segmental glomerulosclerosis. Furthermore, concerns regarding kidney donations by living donors may be alleviated because of the reduced risk of recurrence [28]. In addition, genetic diagnosis aids in choosing potential donors among living relatives. For example, if Alport syndrome is diagnosed in a male patient, his mother needs to be tested for the presence of pathogenic variants before considering donating a kidney to her son. This is of clinical importance because we genetically identified Alport syndrome in patients with a clinical diagnosis of focal segmental glomerulosclerosis. This finding is an indication of variable phenotypic expressions caused by mutations in Col4A genes, such as hematuria or proteinuria, and is consistent with recent studies [29-31].
The limitations of our study include a relatively short study period and small sample size, in addition to its single-center design. Another limitation is that only one patient had a family history of CKD. In addition, some compound heterozygous variants were not investigated for phasing. The significance of variants of uncertain significance was not investigated further because it was beyond the scope of this study. The inherent shortcomings of WES include the possibility of missing mutations in introns, copy number variations, trinucleotide repeat expansions, methylation abnormalities, and mutations in exons with low coverage [23].
In conclusion, WES provided a genetic diagnosis in a considerable proportion of patients with pediatric CKD in Republic of Korea, which may confirm clinical diagnoses, provide guidelines for patient management, and aid in genetic counseling. Our study provides more evidence supporting WES as a new diagnostic method for identifying CKD etiology in children.
Notes
Ethical statements
This study was approved by the Institutional Review Board of Seoul National University Hospital (No. 2011-048-1171). All patients were enrolled after informed consent was obtained from them and their caregivers.
Conflicts of interest
No potential conflict of interest relevant to this article was reported.
Funding
This study was supported by a National Research Foundation of Korea grant funded by the Korean government (MSIT, No. 2020R1A2C1100974).
Author contributions
Conceptualization: YHA, HGK
Data curation: JM, YHA
Formal analysis: HL
Funding acquisition: HGK
Investigation: YHA
Methodology: JM
Project administration: HGK
Visualization: YHA
Writing-original draft: HL
Writing-review & editing: HGK
All authors read and approved the final manuscript.


